Lentivirus Pseudotyped with Coronavirus Spike (S) Protein
To understand the mechanism of coronavirus cell entry and how the viral tropism evolves over time to allow a virus to jump from one host species to another, it is essential to study how S proteins from different coronavirus species interact with their host receptors. However, such studies are hampered by the difficulty to produce and manipulate live coronavirus, especially in cases of dangerous virus strains such as SARS-CoV-2 that require biosafety level 3 (BSL-3) labs.
An alternative to using live coronavirus is to use recombinant lentivirus pseudotyped with the coronavirus S protein. Lentivirus, like coronavirus, is an enveloped virus. When packaging recombinant lentivirus, the S protein could be introduced onto the viral envelope. The resulting pseudotyped lentivirus can now utilize the S protein displayed on its surface to gain entry into host cells possessing the proper receptor, mimicking the mechanism of cell entry by coronavirus. Recombinant lentivirus is very safe and can be pseudotyped with either wildtype or mutant S proteins from any type of coronavirus.
VectorBuilder offers a variety of lentivirus vectors pseudotyped with S proteins from a wide range of coronavirus species. Different vectors can be used to express different reporters such as EGFP or luciferase, allowing the monitoring of viral entry into host cells by a variety of means.
Service Details
- Price and turnaround
- Types of lentivirus vectors used in pseudotyping
- Types of spike (S) proteinsused in pseudotyping
Technical Information
- Mechanism of coronavirus cell entry
- Experimental validation
- Documents
How to Order
Service Details
Price and turnaround Price Match
| Scale | Application | Minimum Titer | Volume | Price (USD) | Turnaround |
|---|---|---|---|---|---|
| Pilot | Cell culture | >108 TU/ml | 250 ul (10x25 ul) | $899 |
6-12 days
|
| Medium | 1 ml (10x100 ul) | $1,299 | |||
| Large | >109 TU/ml | 1 ml (10x100 ul) | $2,199 | ||
| Ultra-purified medium | Cell culture & in vivo | >109 TU/ml | 500 ul (10x50 ul) | $2,799 | |
| Ultra-purified large | 1 ml (10x100 ul) | $3,399 |
TU = Transduction units (also known as infectious unit)
Types of lentivirus vectors used in pseudotyping
In theory, any lentivirus vector can be pseudotyped. For your convenience, we provide a set of standard vectors that could satisfy most applications. You can use the vector picker below to find a standard vector suitable for you:
Vector Picker
Map of lentivirus vector used in pseudotyping
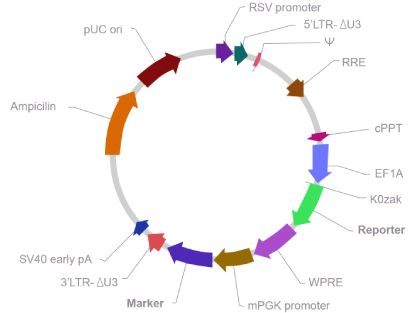
If you can’t a find a suitable vector above, you can create a custom lentivirus vector as follows:
Click here to design a vector using our online design tool
Click here to send us a vector design request
Types of spike (S) proteins used in pseudotyping
Coronavirus constitutes a vast group of viruses that are extremely widespread in nature, infecting virtually all mammals and birds examined. Hundreds of coronavirus species have been characterized thus far. Of these, a few dozen can be deemed important because they infect humans, livestock, pets, or model animals, or they are evolutionarily close related to them. The phylogenetic tree of these import coronavirus species is shown below:
Phylogenetic Tree of Important Coronavirus Species
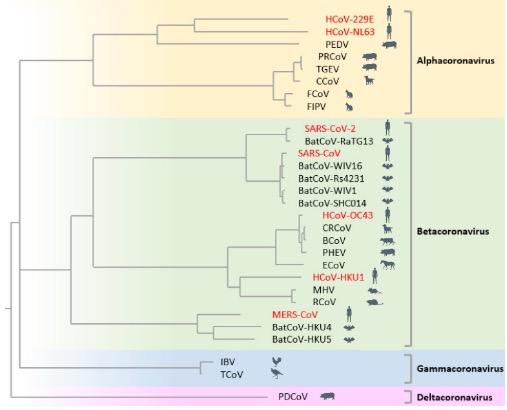
More detailed information of these important coronavirus species is listed in the table below:
List of important coronavirus species
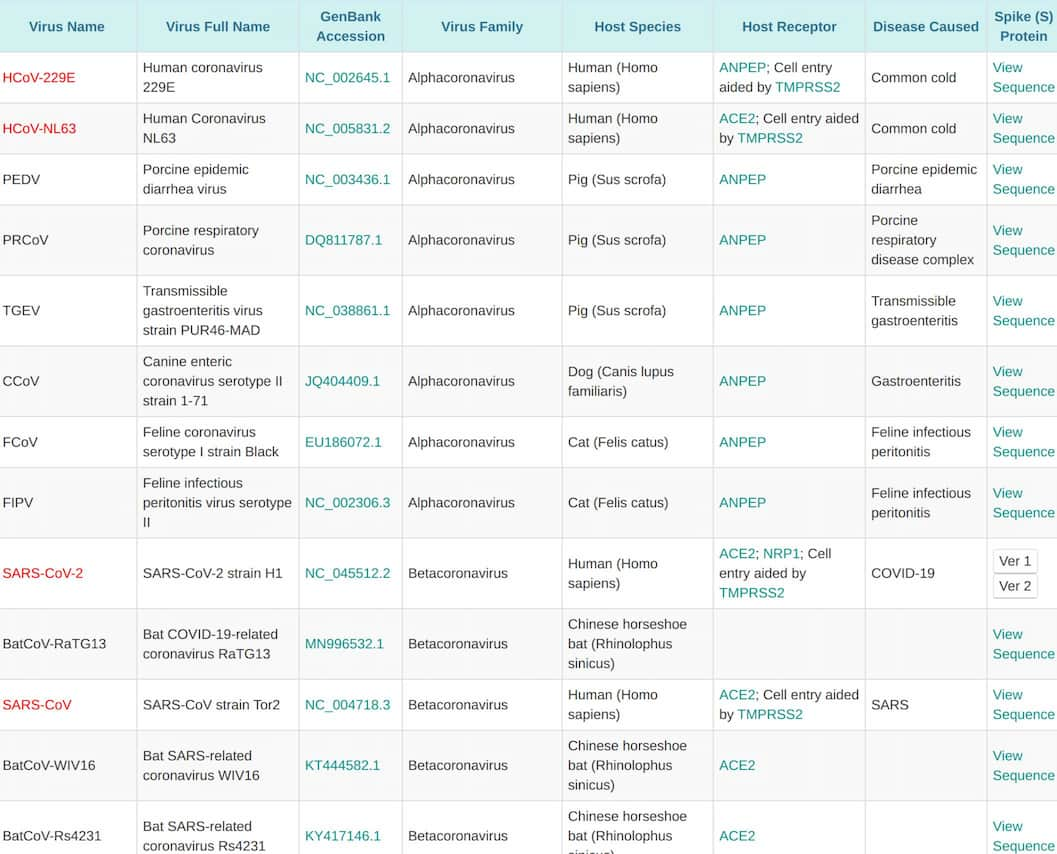
VectorBuilder offers lentivirus pseudotyped by S proteins of any of the above coronavirus species. Pseudotyping by other sources of S proteins can also be requested.
For pseudotyping with the SARS-Cov-2 S protein, we provide several options as listed in the table below:
List of currently offered
SARS-CoV-2 S protein variants
VectorBuilder also offers bald lentivirus lacking viral envelope protein, which can be used as negative control.
Technical Information
Mechanism of coronavirus cell entry
The first step for viruses to gain entry into host cells is to attach themselves onto the cell surface. This is achieved when specific receptor proteins on the virus bind to complementary receptors on the host cell membrane. In the case of enveloped viruses such as coronavirus and lentivirus, which have a lipid bilayer membrane as their outer layer, the viral receptor proteins are embedded in the envelope and protrude outward. In the case of non-enveloped viruses such as adenovirus and AAV, which have a protein capsid shell as their outer layer, the viral receptors are a part of the capsid.
For coronavirus, the spike (S) protein forms the spikes protruding out of the viral envelope, giving the virus a crown or halo-like appearance – hence the name coronavirus. These spikes are responsible for viral attachment to host cells, and they also mediate subsequent fusion of the viral envelope to the host cell membrane leading to viral entry into cells.
Coronavirus constitutes a large group of viruses that display very different tropisms for different host species. As such, a given coronavirus species generally can only infect one or a few host species effectively. This tropism is due to the specific binding of the S protein from a given type of virus to its target receptor of the host species. For example, SARS-CoV and SARS-CoV-2, the viruses responsible for SARS and COVID-19, respectively, infect humans via the binding of their S protein to the angiotensin converting enzyme 2 (ACE2) receptor on the surface of human respiratory, lung and oral epithelial cells. On the other hand, MERS-CoV, which is responsible for MERS, gains entry into human cells via binding to the dipeptidyl peptidase-4 (DPP4) receptor.
Experimental Validation
We developed a number of proprietary techniques to optimize our pseudotyping protocol. As a result, our optimized virus exhibits much higher transduction efficiency compared to published protocols. We carried out extensive experimental validation of our virus. Our results showed that lentivirus pseudotyped with SARS-CoV-2 wildtype S protein or its variants could efficiently transduce 293T cells expressing high levels of the human ACE2 receptor, but could not transduce 293T cells lacking ACE2 expression (Figure 1). We, therefore, recommend using our ACE2-expressing cell lines that have been optimized for transduction with SARS-CoV-2 S protein or VSV-G pseudotyped virus.
Below are the results of transduction test (Figure 1, Figure 2, Figure 3), luciferase (Figure 4) and Western blot (Figure 5).

Figure 1. 293T or human ACE2 receptor (hACE2) expressing 293T cells were transduced with EGFP expressing lentivirus pseudotyped with SARS-CoV-2 wildtype (WT), B.1.1.529 (omicron), or D614G S protein. Bald lentivirus was used as a negative control. Images were taken 72 h post transduction.
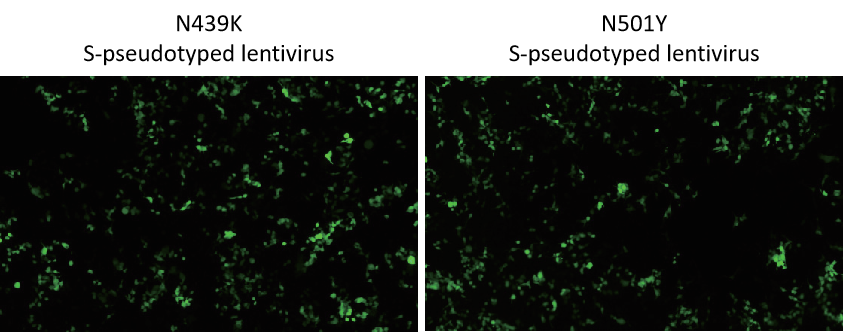
Figure 2. 293T (hACE2) cells transduced with EGFP expressing lentivirus pseudotyped with N439K or N501Y S protein.
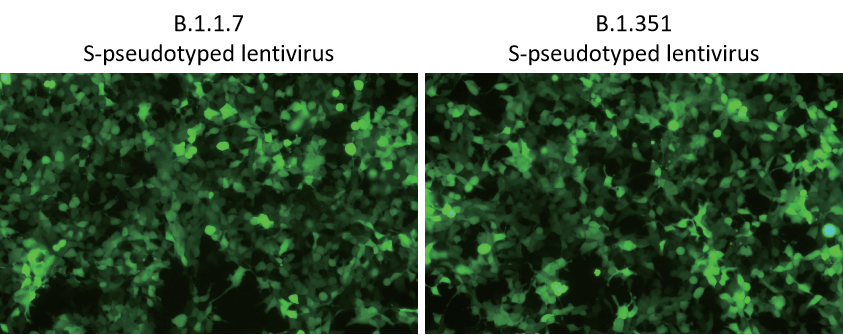
Figure 3. 293T (hACE2) cells transduced with EGFP expressing lentivirus pseudotyped with B.1.1.7 or B.1.351 S protein

Figure 4. 293T (hACE2) cells were transduced with luciferase expressing lentivirus pseudotyped with SARS-CoV-2 wildtype S protein or its D614G variant or VSV-G protein. Luciferase assay was performed 72 h post transduction and the background luminescence from untransduced cells was subtracted from the luminescence value of each group.
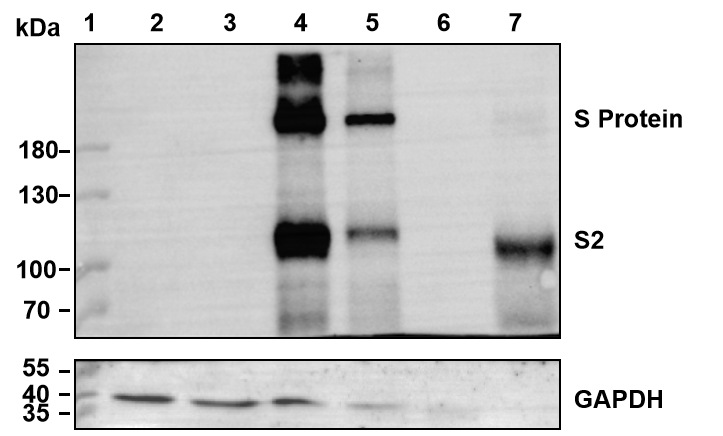
Figure 5. Western blot analysis using an antibody against the S2 domain of the SARS-CoV-2 S protein revealed the presence of both the full-length and the cleaved S2 portion of S in packaging cells and in pseudotyped virus. Note: the S protein naturally undergoes cleavage to facilitate viral entry into infected cells. Lane 1: marker. Lane 2: Untransduced 293T cells. Lane 3: 293T cells transfected with VSV-G pseudotyping lentivirus packaging plasmids. Lane 4: 293T cells transfected with S protein helper plasmid alone. Lane 5: 293T cells transfected with S pseudotyping lentivirus packaging plasmids. Lane 6: VSV-G pseudotyped lentiviral particles. Lane 7: S pseudotyped lentiviral particles.
How to Order
You can inquire about our lentivirus pseudotyping services by following the link below:
Click here to send us a lentivirus pseudotyping request


